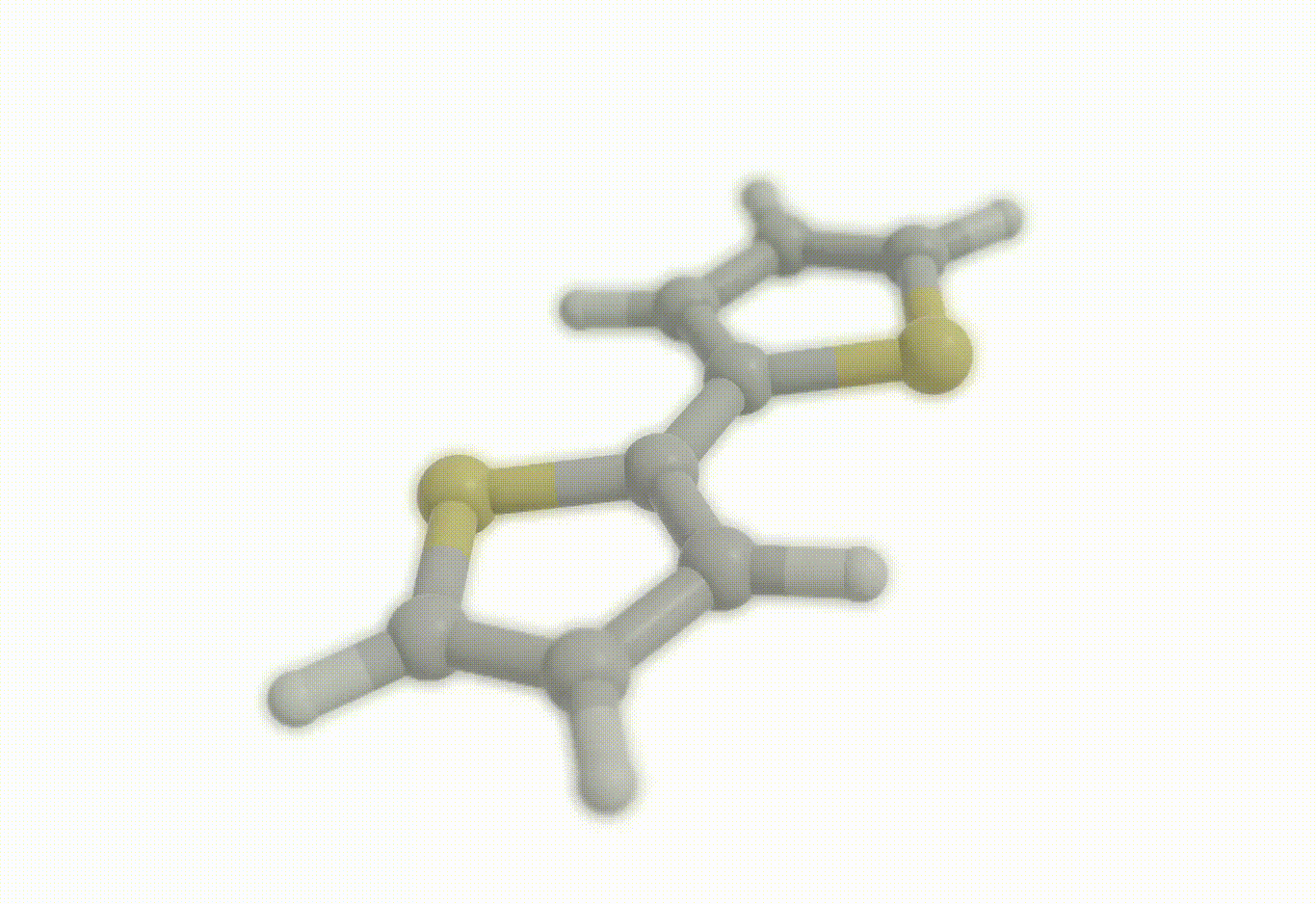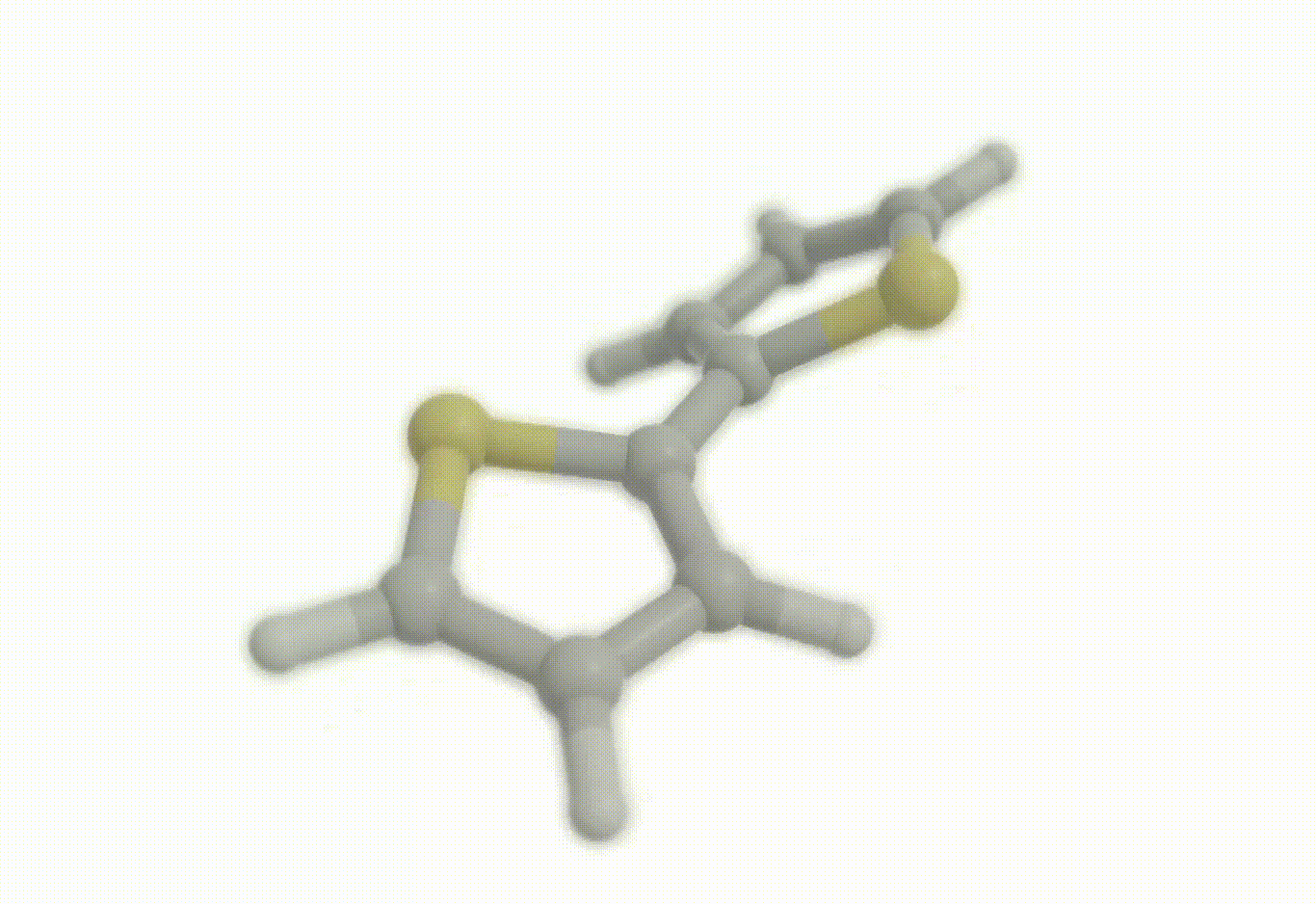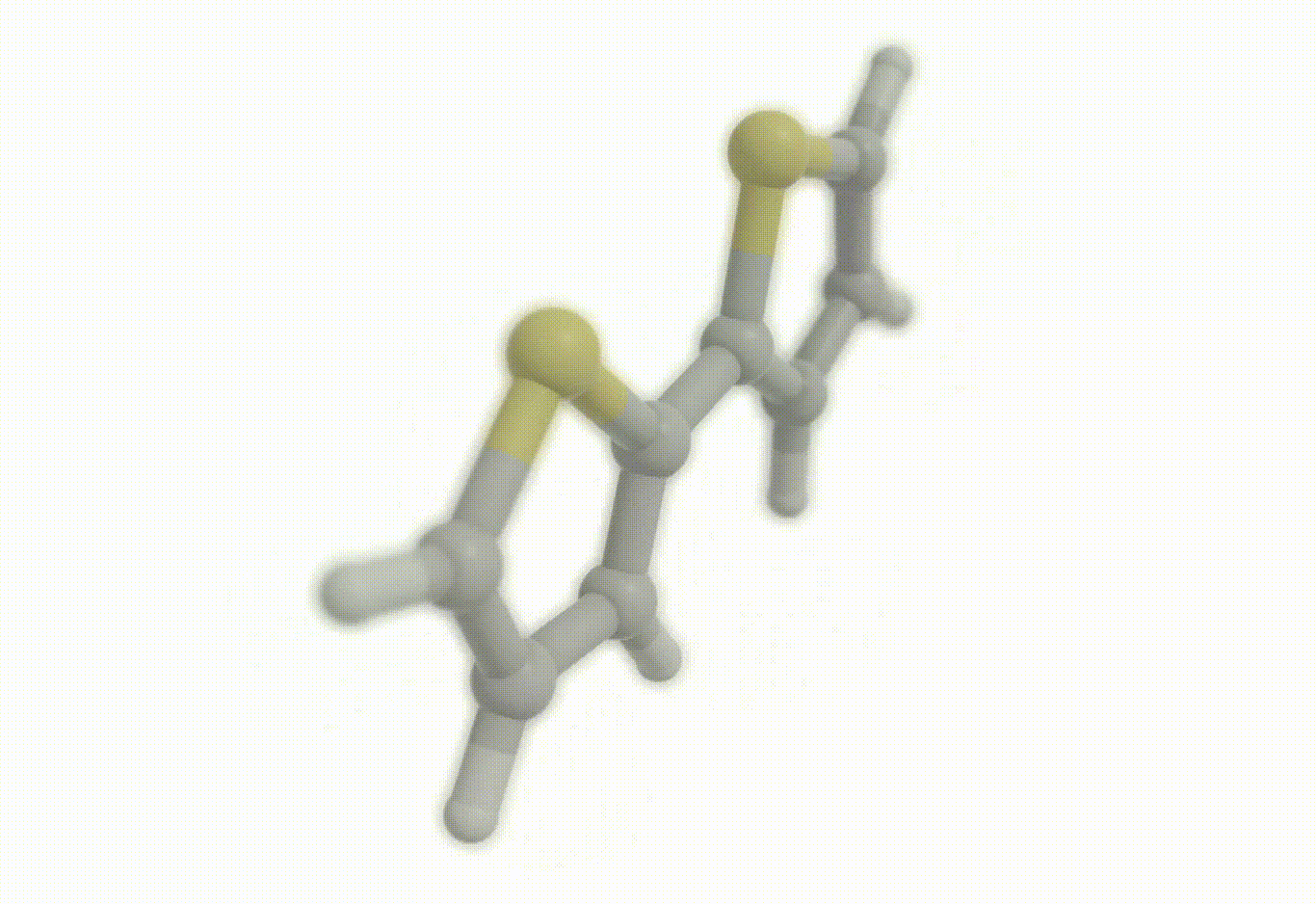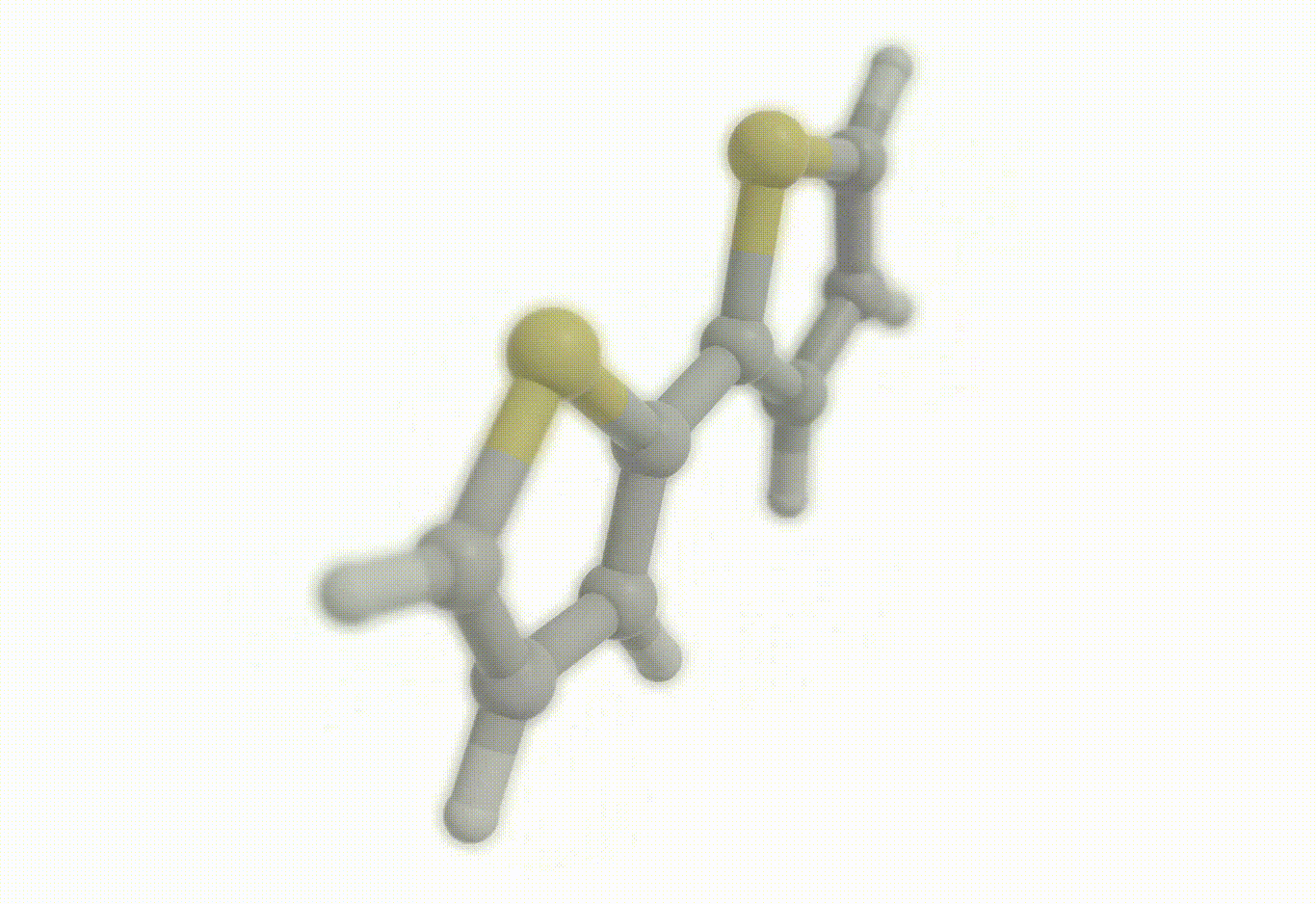
Potential energy surfaces#
Structure optimizations are performed with the aid of geomeTRIC [WS16].
Ground state optimization#
Python script
import veloxchem as vlx
xyz="""
...
"""
molecule = vlx.Molecule.read_xyz_string(xyz)
basis = vlx.MolecularBasis.read(molecule, 'def2-svp')
scf_drv = vlx.ScfRestrictedDriver()
scf_drv.filename = 'mol-opt'
scf_drv.xcfun = 'b3lyp'
scf_drv.dispersion = True
results = scf_drv.compute(molecule, basis)
opt_drv = vlx.OptimizationDriver(scf_drv)
opt_drv.filename = 'mol-opt'
opt_results = opt_drv.compute(molecule, basis, results)
Download a Python script type of input file to perform an optimization for the bithiophene molecule at the B3LYP+D4/def2-svp level of theory.
Text file
Please refer to the keyword list for a complete set of options. Dispersion can be activated in the @method settings section by using the keyword dispersion.
@jobs
task: optimize
@end
@method settings
xcfun: b3lyp
basis: def2-svp
dispersion: yes # use dft-d4 correction
@end
@molecule
charge: 0
multiplicity: 1
xyz:
...
@end
Download a text file type of input file to perform an optimization for the bithiophene molecule at the B3LYP+D4/def2-svp level of theory.
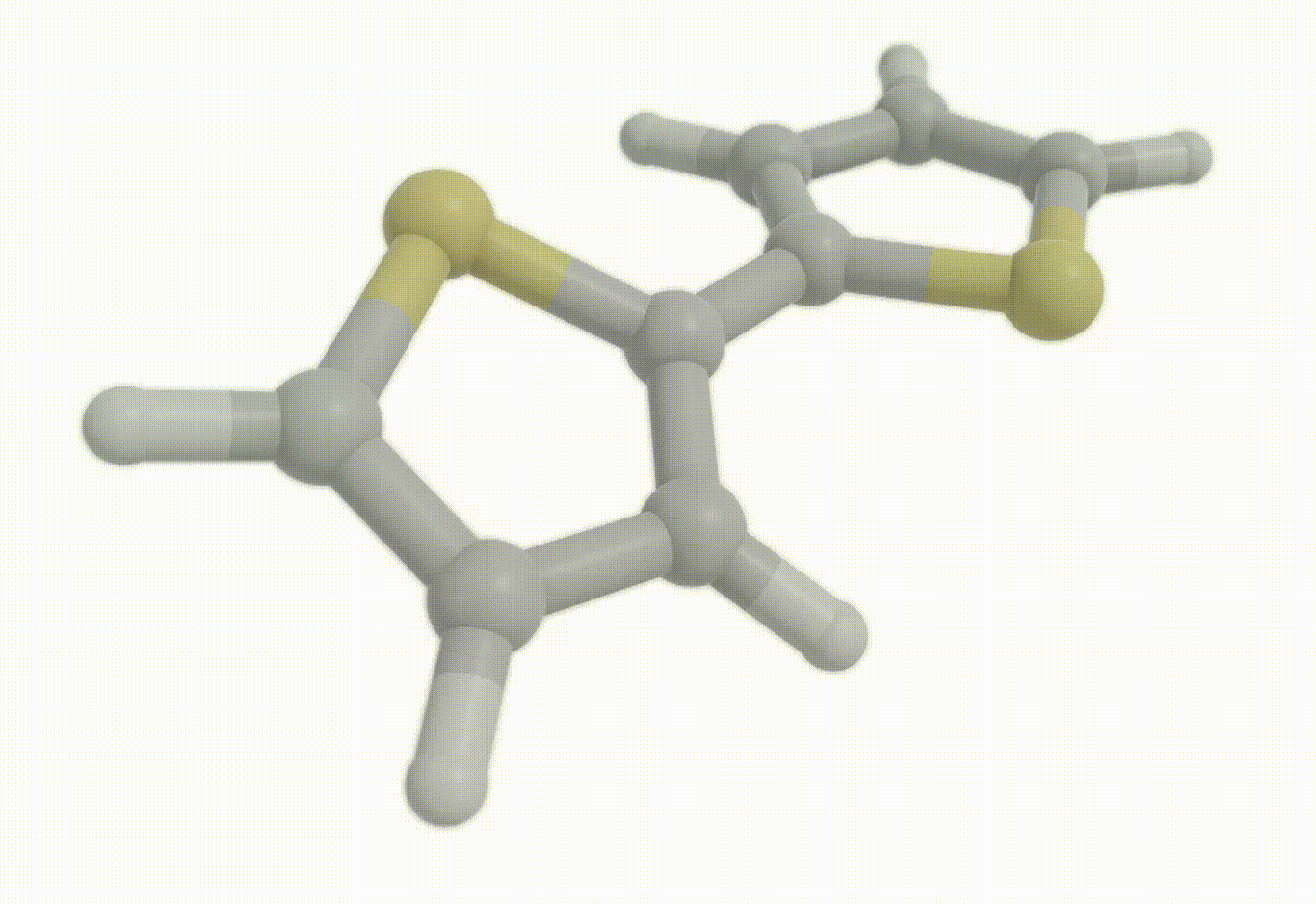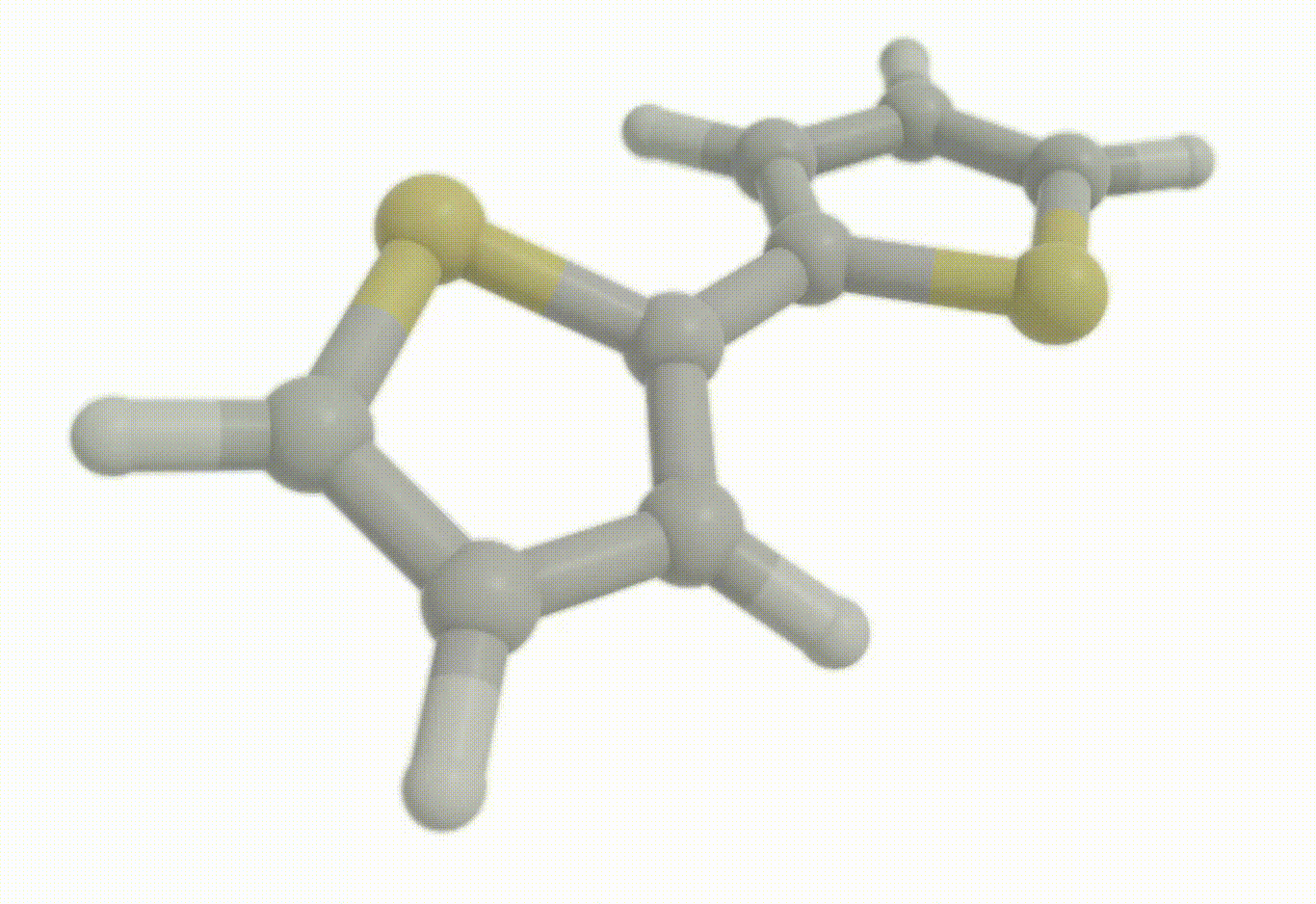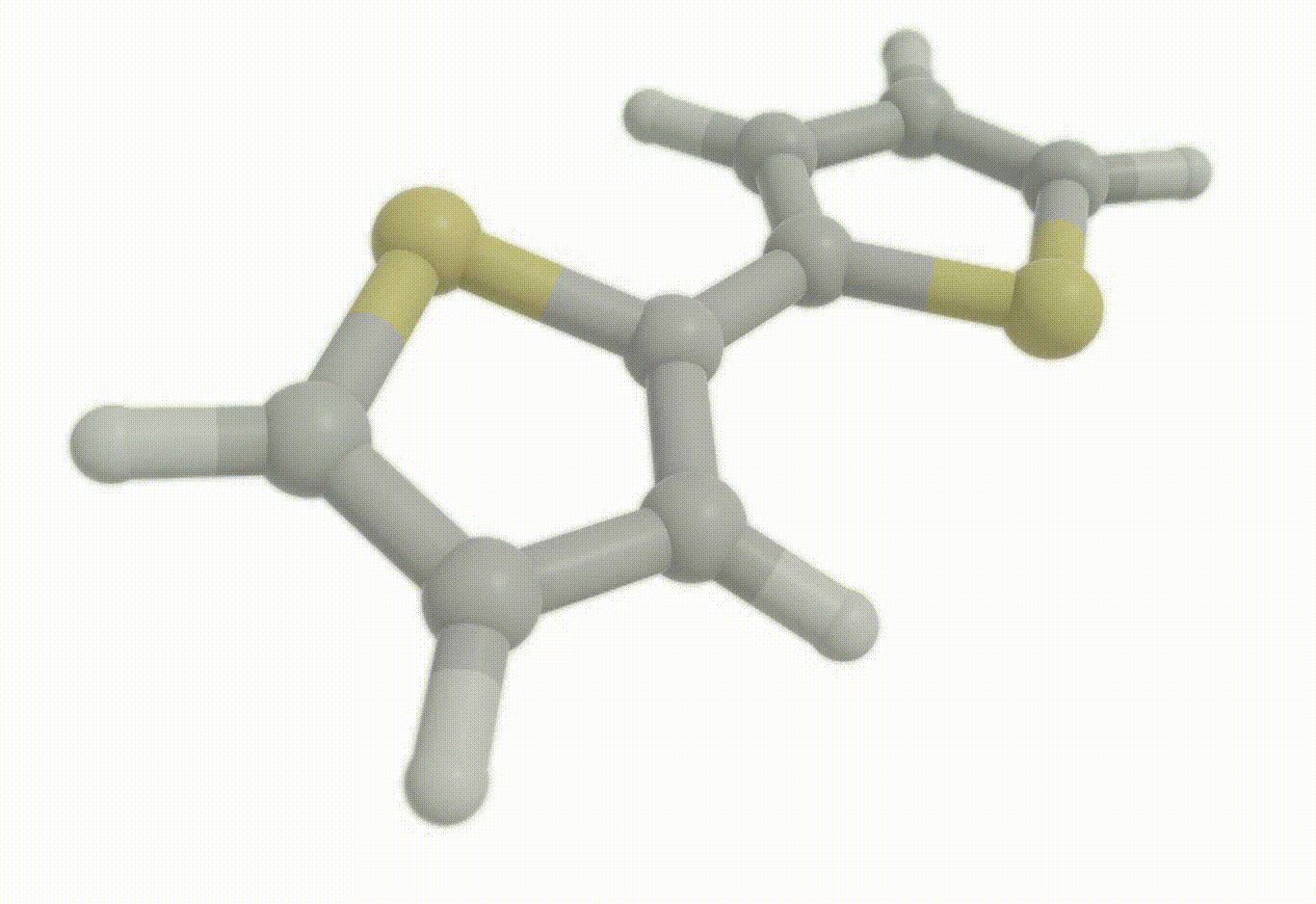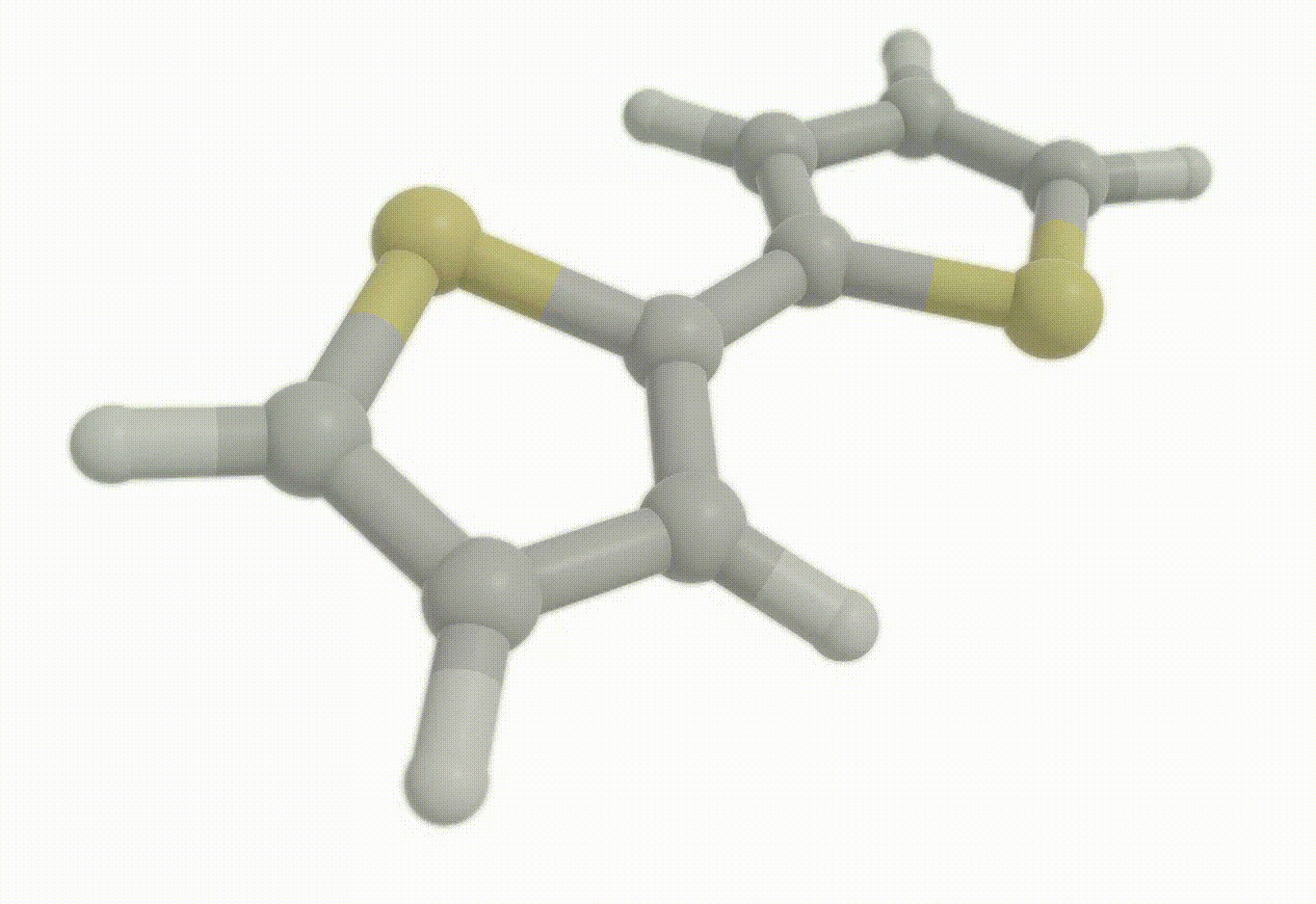
Excited state optimization#
Python script
import veloxchem as vlx
xyz="""
...
"""
molecule = vlx.Molecule.read_xyz_string(xyz)
basis = vlx.MolecularBasis.read(molecule, 'def2-svp')
scf_drv = vlx.ScfRestrictedDriver()
scf_drv.filename = 'mol-opt'
scf_drv.xcfun = 'b3lyp'
scf_drv.dispersion = True
scf_results = scf_drv.compute(molecule, basis)
rsp_drv = vlx.LinearResponseEigenSolver()
rsp_drv.nstates = 2
rsp_results = rsp_drv.compute(molecule, basis, scf_results)
grad_drv = vlx.TddftGradientDriver(scf_drv)
grad_drv.state_deriv_index = 1
opt_drv = vlx.OptimizationDriver(grad_drv)
opt_drv.filename = 'mol-S1-opt'
opt_results = opt_drv.compute(molecule, basis, scf_drv, rsp_drv, rsp_results)
Download a Python script type of input file to perform an optimization of the first excited state of bithiophene molecule at the B3LYP/def2-svp level of theory.
Text file
To optimize an excited state, you need to use the task optimize and to specify which state you want to optimize with the state_deriv_index: keyword in the @gradient section.
@jobs
task: optimize
@end
@response
property: absorption
nstates: 2
@end
@gradient
state_deriv_index: 1
@end
@method settings
xcfun: b3lyp
basis: def2-svp
@end
@molecule
charge: 0
multiplicity: 1
xyz:
...
@end
Download a text file type of input file to perform an optimization of the first excited state of bithiophene molecule at the B3LYP/def2-svp level of theory.
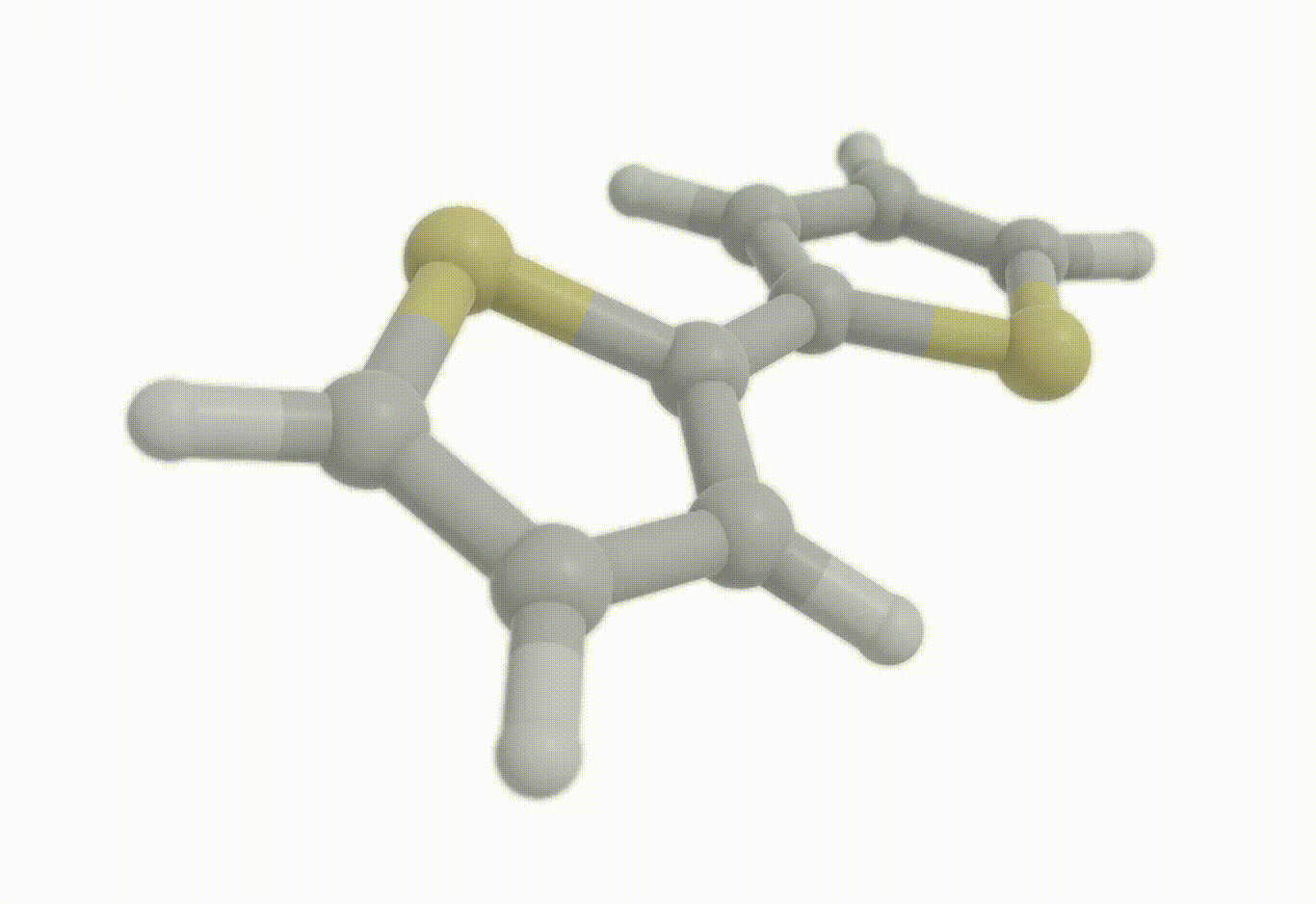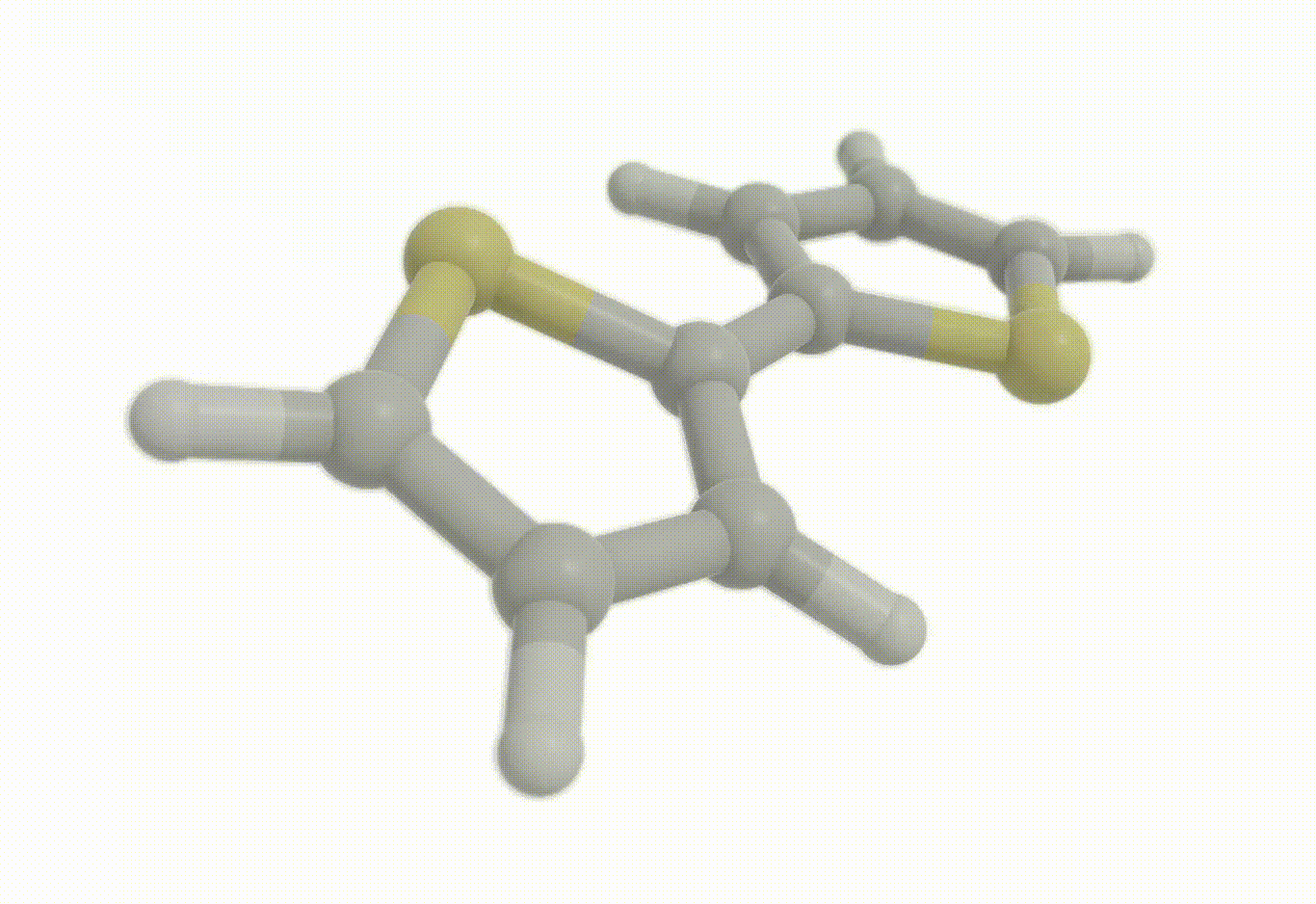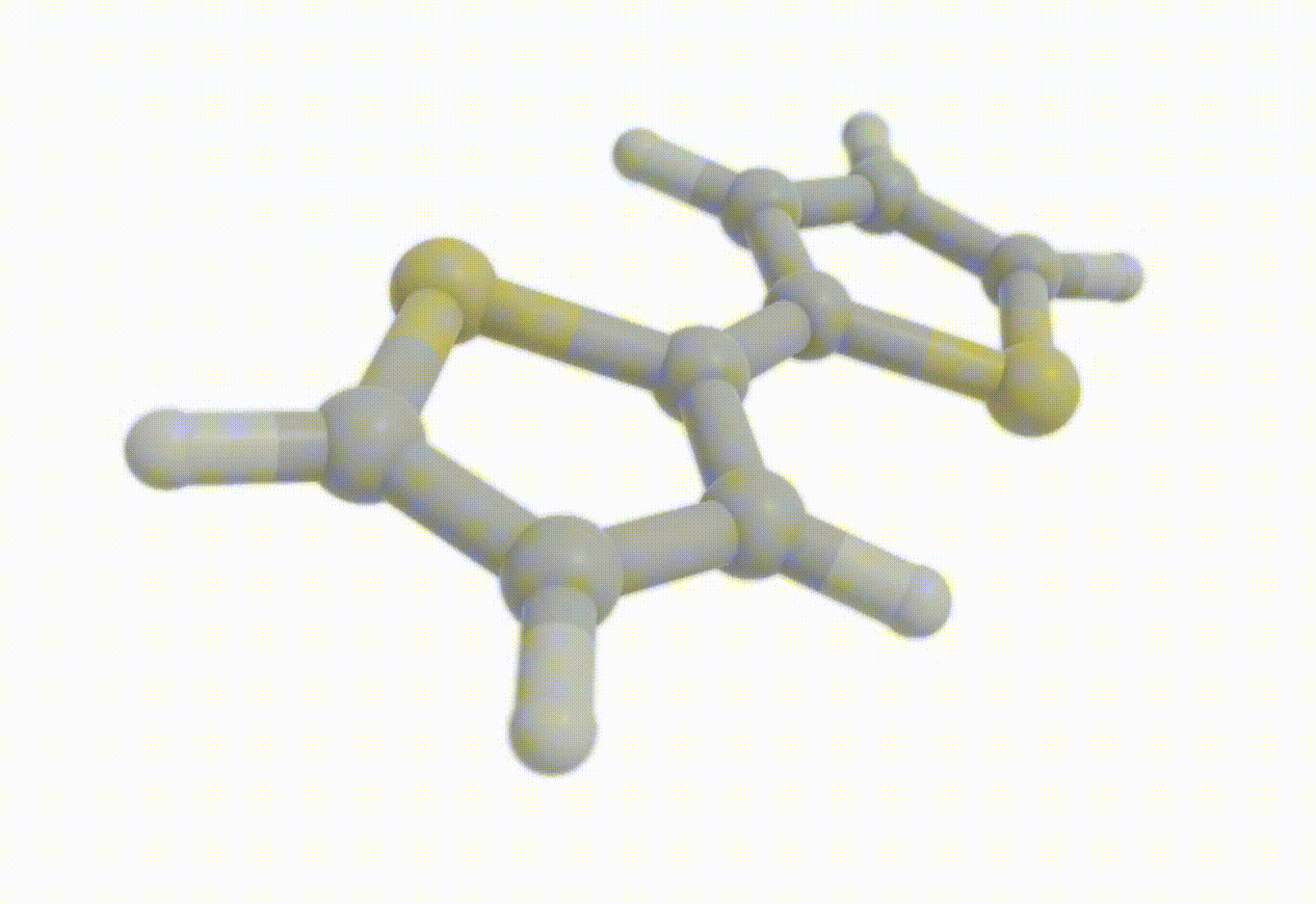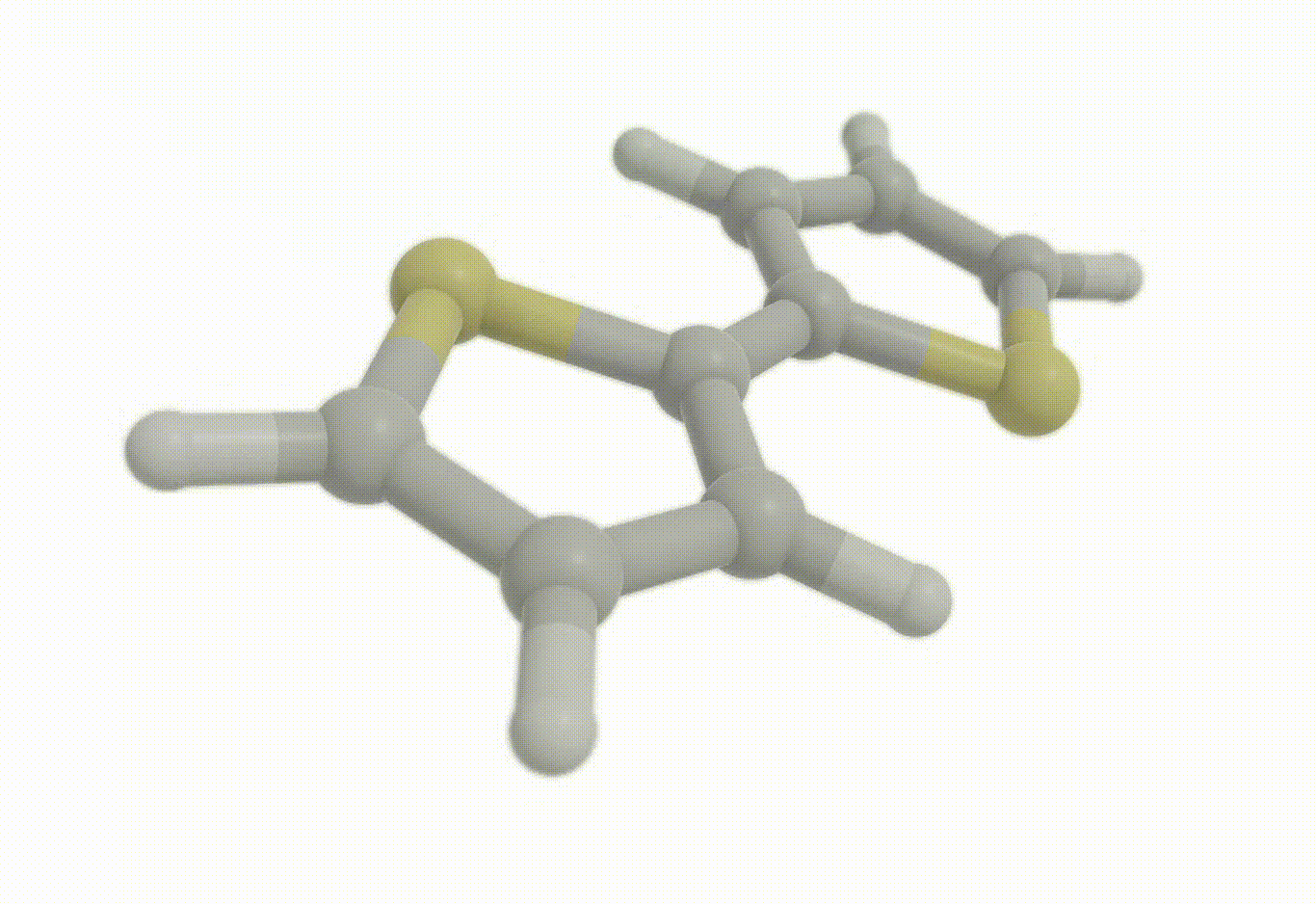
Constrained optimization#
Internal coordinates (distances, angles, dihedrals) can be constrained during the molecular structure optimization with use of either the set, freeze, or scan options.
Set or freeze internal coordinate#
set will aim at converging an internal coordinate to a desired value while freeze will keep it at its initial value.
Python script
import veloxchem as vlx
xyz="""
...
"""
molecule = vlx.Molecule.read_xyz_string(xyz)
basis = vlx.MolecularBasis.read(molecule, 'def2-svp')
scf_drv = vlx.ScfRestrictedDriver()
scf_drv.filename = 'bithio-freeze'
scf_drv.xcfun = 'b3lyp'
results = scf_drv.compute(molecule, basis)
opt_drv = vlx.OptimizationDriver(scf_drv)
opt_drv.constraints = [ "set dihedral 1 3 4 2 90.0", "freeze distance 3 4"]
opt_drv.filename = 'bithio-freeze'
opt_results = opt_drv.compute(molecule, basis, results)
Download a Python script type of input file to perform a optimization of the bithiophene molecule at the B3LYP/def2-svp level of theory where the inter-ring distance has been frozen to the initial geometry and where the inter-ring dihedral (S-C-C-S) has been constrained to 90°.
Text file
@jobs
task: optimize
@end
@method settings
xcfun: b3lyp
basis: def2-svp
@end
@optimize
set dihedral 1 3 4 2 90.0
freeze distance 3 4
@end
@molecule
charge: 0
multiplicity: 1
xyz:
...
@end
Download a text file type of input file to perform a optimization of the bithiophene molecule at the B3LYP/def2-svp level of theory where the inter-ring distance has been frozen to the initial geometry and where the inter-ring dihedral (S-C-C-S) has been constrained to 90°.
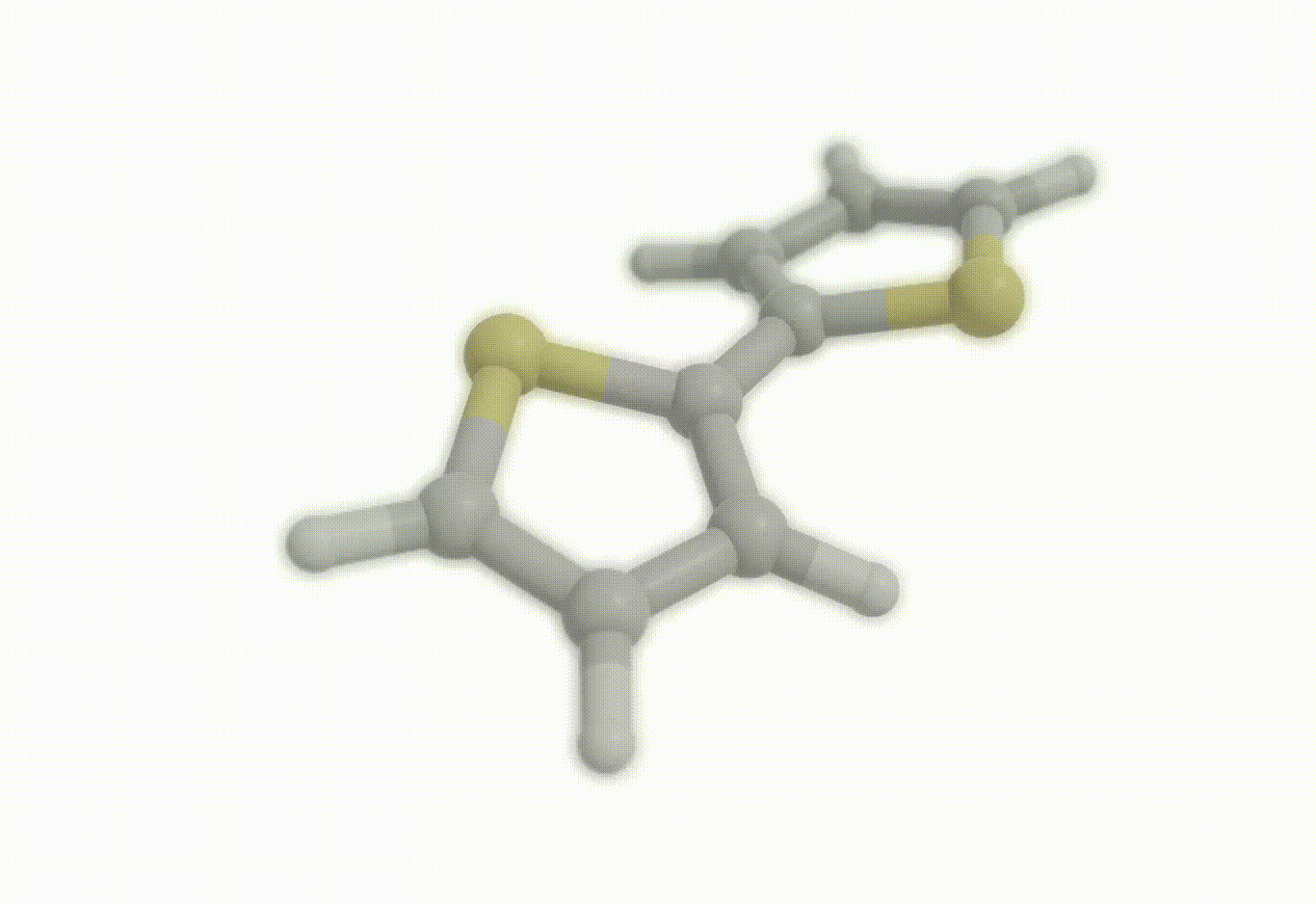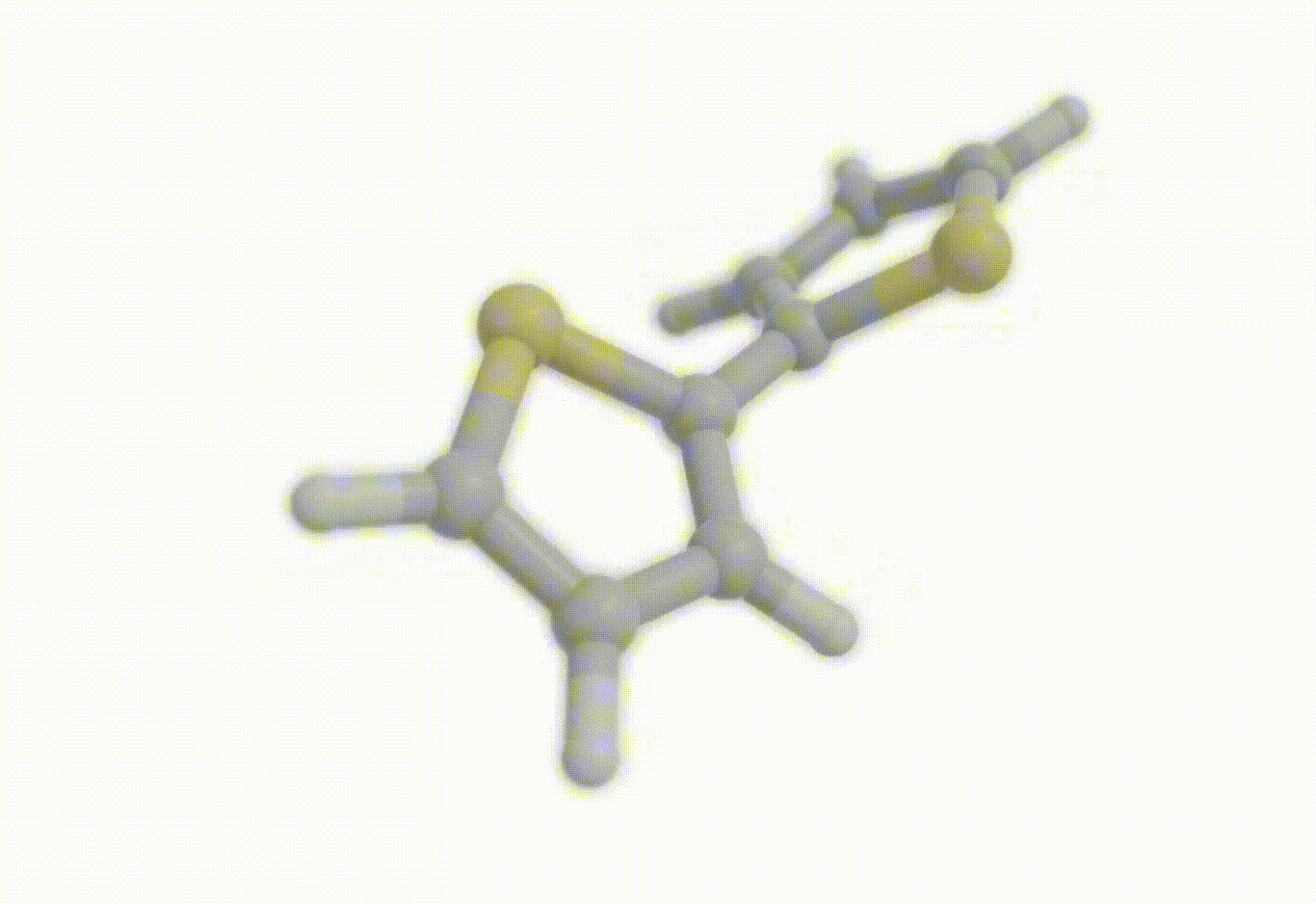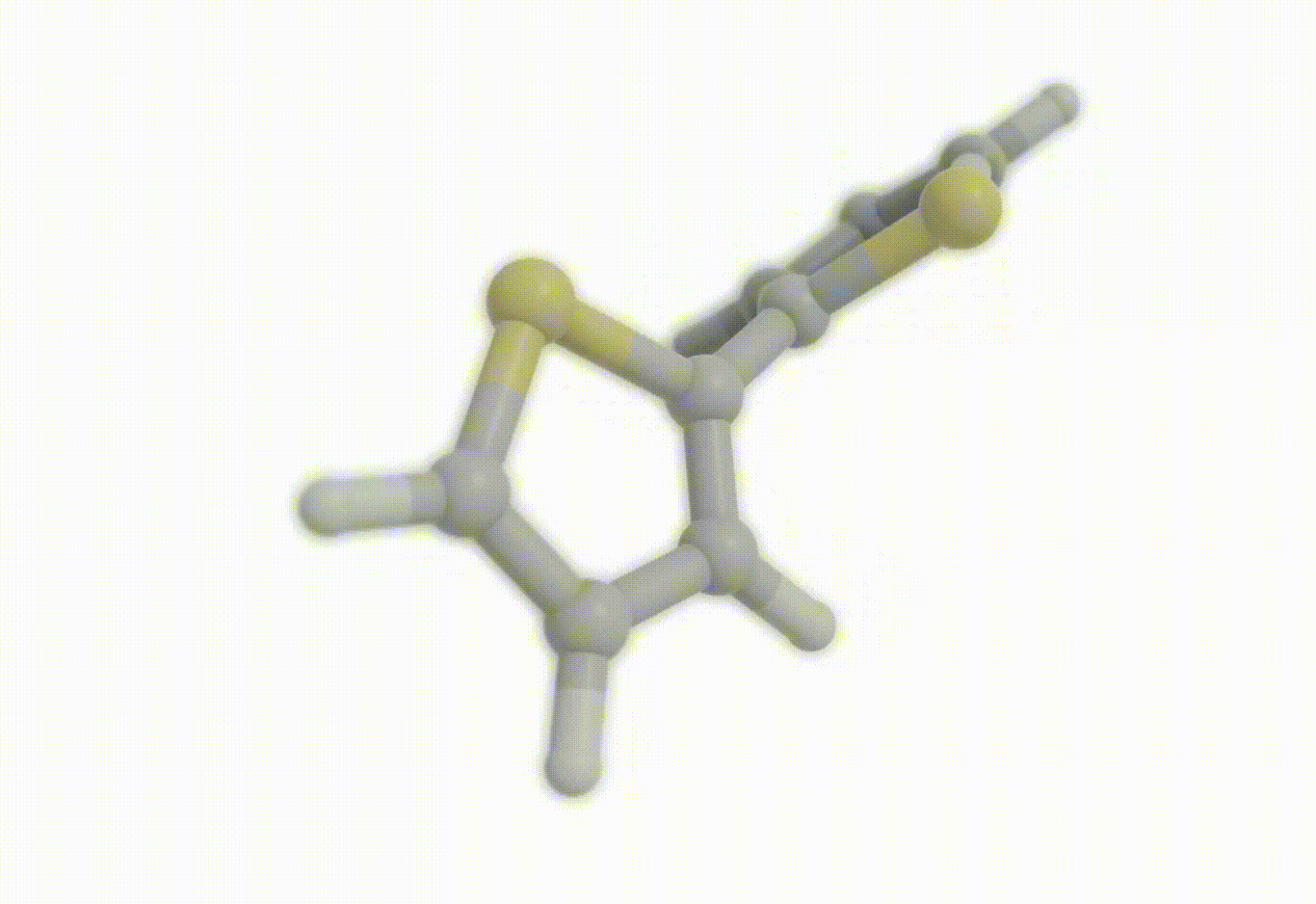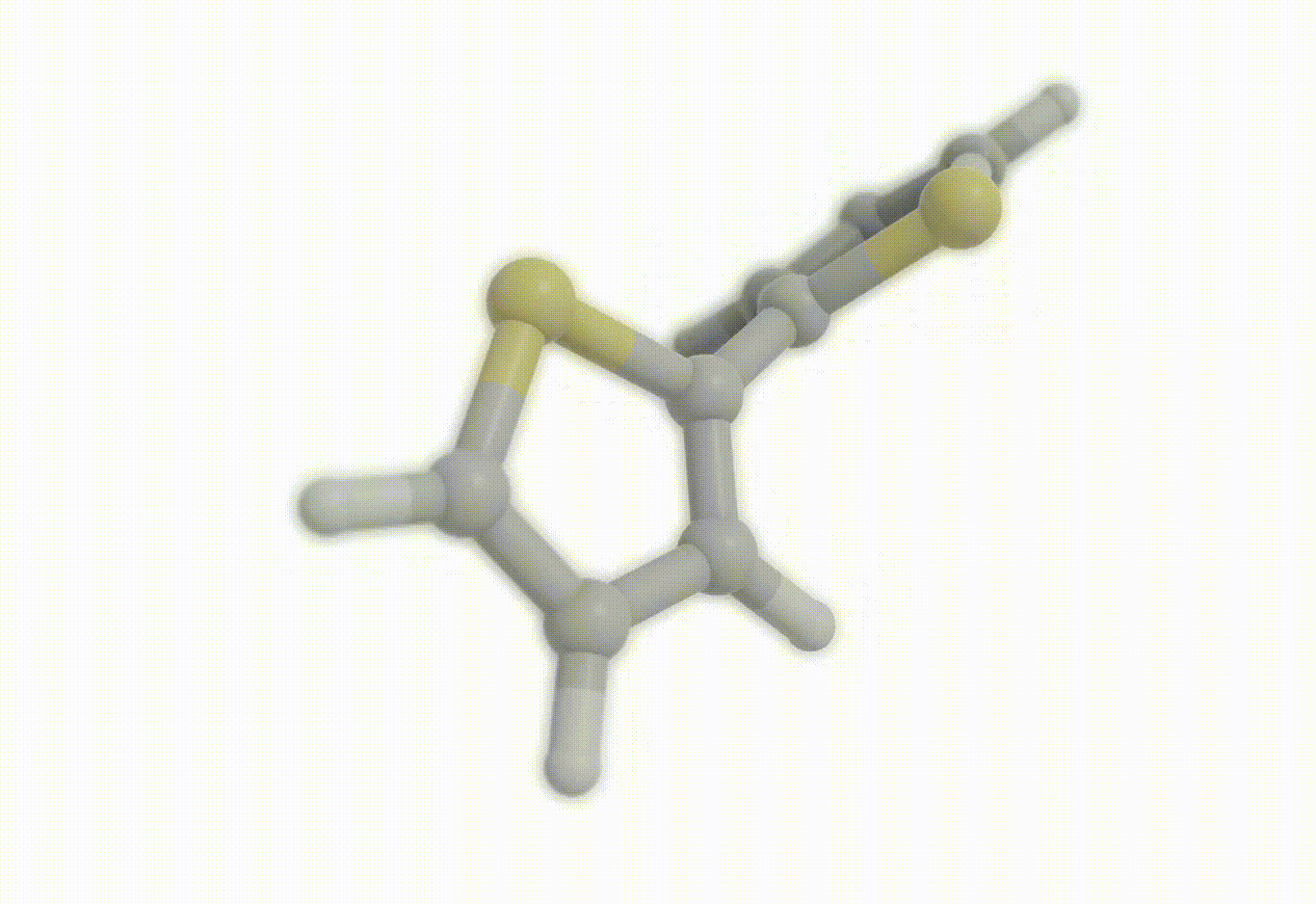
Scan coordinate#
scan will scan an internal coordinate from an initial to a final value in a given number of steps.
scan distance 6 1 1.4 1.5 9
scan angle 6 1 2 100 110 9
scan dihedral 6 1 2 3 0 360 19
Python script
import veloxchem as vlx
xyz="""
...
"""
molecule = vlx.Molecule.read_xyz_string(xyz)
basis = vlx.MolecularBasis.read(molecule, 'def2-svp')
scf_drv = vlx.ScfRestrictedDriver()
scf_drv.filename = "bithio-scan"
scf_drv.xcfun = "b3lyp"
results = scf_drv.compute(molecule, basis)
opt_drv = vlx.OptimizationDriver(scf_drv)
opt_drv.constraints = ["scan dihedral 1 3 4 2 180 0 9"]
opt_drv.filename = "bithio-scan"
opt_results = opt_drv.compute(molecule, basis, results)
Download a Python script type of input file to perform a relaxed scan of the bithiophene molecule at the B3LYP/def2-svp level of theory where the inter-ring dihedral (S-C-C-S) is scanned from 180° to 0° in 9 steps.
Text file
@jobs
task: optimize
@end
@method settings
xcfun: b3lyp
basis: def2-svp
@end
@optimize
scan dihedral 1 3 4 2 180 0 9
@end
@molecule
charge: 0
multiplicity: 1
xyz:
...
@end
Download a text file type of input file to perform a relaxed scan of the bithiophene molecule at the B3LYP/def2-svp level of theory where the inter-ring dihedral (S-C-C-S) is scanned from 180° to 0° in 9 steps.