Program structure#
import veloxchem as vlx
The VeloxChem program adopts a layered Python/C++ language structure.
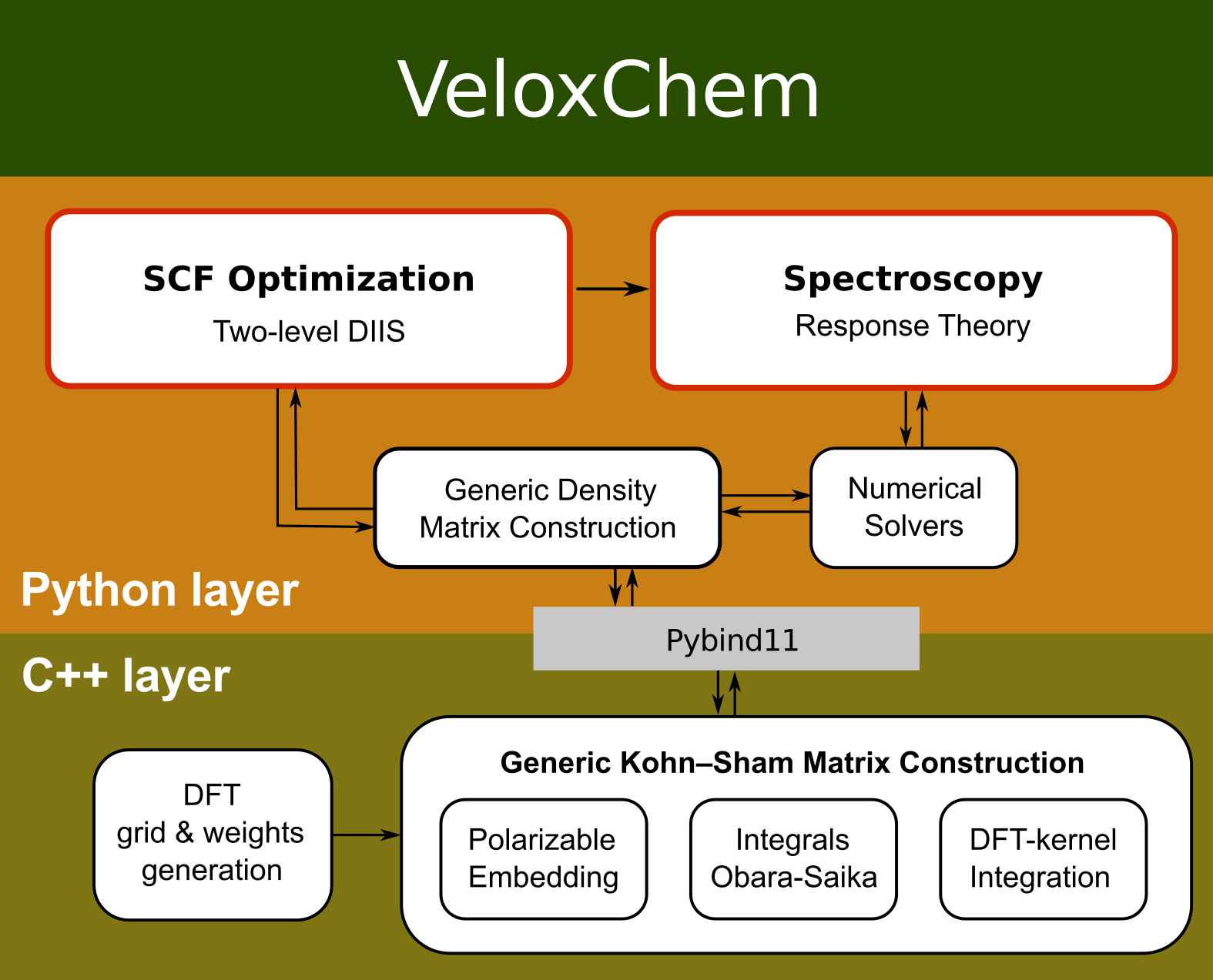
You can use the standard functions to get a more detailed view of the Python layer. The help function will give you a package description, stating the copyright agreement.
help(vlx)
Help on package veloxchem:
NAME
veloxchem
DESCRIPTION
# VELOXCHEM
# ----------------------------------------------------
# An Electronic Structure Code
#
# SPDX-License-Identifier: BSD-3-Clause
#
# Copyright 2018-2025 VeloxChem developers
#
# Redistribution and use in source and binary forms, with or without modification,
# are permitted provided that the following conditions are met:
#
# 1. Redistributions of source code must retain the above copyright notice, this
# list of conditions and the following disclaimer.
# 2. Redistributions in binary form must reproduce the above copyright notice,
# this list of conditions and the following disclaimer in the documentation
# and/or other materials provided with the distribution.
# 3. Neither the name of the copyright holder nor the names of its contributors
# may be used to endorse or promote products derived from this software without
# specific prior written permission.
#
# THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND
# ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED
# WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
# DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
# FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
# DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
# SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION)
# HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT
# LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT
# OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
PACKAGE CONTENTS
__main__
aodensitymatrix
aoindices
atombdedriver
atomtypeidentifier
batchsize
blockdavidson
c2diis
c6driver
checkpoint
cli
conformergenerator
cpcmdriver
cphfsolver
cppsolver
cubicgrid
cubicresponsedriver
densityviewer
densityvieweralternative
dftutils
dispersionmodel
distributedarray
doubleresbeta
embedding
environment
errorhandler
espchargesdriver
evbdataprocessing
evbdriver
evbfepdriver
evbreporter
evbsystembuilder
excitedstateanalysisdriver
excitondriver
features
firstorderprop
fockdriver
gradientdriver
griddriver
hessiandriver
hessianorbitalresponse
imdatabasedriver
imdatabasepointcollecter
imforcefieldgenerator
impescoordinates
impesdriver
inputparser
interpolationdatapoint
interpolationdriver
linearsolver
lreigensolver
lreigensolverunrest
lrsolver
lrsolverunrest
main
matrices
matrix
mklconf
mmdriver
mmforcefieldgenerator
mmgradientdriver
mofbuilder
mofmdprepare
mofoptimizer
mofpreparer
mofutils
mointsdriver
molecularbasis
molecularorbitals
molecule
mp2driver
mpitask
nonlinearsolver
numerovdriver
oneeints
openmmdriver
openmmdynamics
openmmgradientdriver
optimizationdriver
optimizationengine
orbitalviewer
outputstream
peforcefieldgenerator
polarizabilitygradient
polorbitalresponse
profiler
pubchemfetcher
pulsedrsp
quadraticresponsedriver
reactionmatcher
reaffbuilder
respchargesdriver
rifockdriver
rigradientdriver
rijkfockdriver
rspabsorption
rspc6
rspcdspec
rspcustomproperty
rspdoublerestrans
rsplinabscross
rsppolarizability
rspproperty
rspshg
rspthreepatransition
rsptpa
rsptpatransition
sadguessdriver
sanitychecks
scfdriver
scfgradientdriver
scfhessiandriver
scfrestdriver
scfrestopendriver
scfunrestdriver
seminario
shgdriver
smddriver
smdsolventproperties
solvationbuilder
solvationfepdriver
subcommunicators
submatrix
symmetryanalyzer
symmetryoperations
tdacppsolver
tdaeigensolver
tdaeigensolverunrest
tddftgradientdriver
tddftorbitalresponse
tdhfhessiandriver
threepatransitiondriver
tmparameters
tpadriver
tpafulldriver
tpareddriver
tpatransitiondriver
trajectorydriver
tsguesser
uffparameters
veloxchemlib
vibrationalanalysis
visualizationdriver
waterparameters
xtbdriver
xtbgradientdriver
xtbhessiandriver
FUNCTIONS
available_functionals(...) method of builtins.PyCapsule instance
available_functionals() -> list[str]
Gets a list of available exchange-correlation functionals.
available_pdft_functionals(...) method of builtins.PyCapsule instance
available_pdft_functionals() -> list[str]
Gets a list of available pdft exchange-correlation functionals components.
bohr_in_angstrom(...) method of builtins.PyCapsule instance
bohr_in_angstrom() -> float
Gets Bohr value in Angstroms.
chemical_element_identifier(...) method of builtins.PyCapsule instance
chemical_element_identifier(arg0: str) -> int
Gets chemical element identifier.
chemical_element_label(...) method of builtins.PyCapsule instance
chemical_element_label(arg0: int) -> str
Gets chemical element label.
chemical_element_mass(...) method of builtins.PyCapsule instance
chemical_element_mass(arg0: int) -> float
Gets chemical element mass.
chemical_element_max_angular_momentum(...) method of builtins.PyCapsule instance
chemical_element_max_angular_momentum(arg0: int) -> int
Gets maximum angular momentum of atomic shell in chemical element.
chemical_element_name(...) method of builtins.PyCapsule instance
chemical_element_name(arg0: int) -> str
Gets chemical element name.
compute_angular_momentum_integrals(...) method of builtins.PyCapsule instance
compute_angular_momentum_integrals(molecule: veloxchem.veloxchemlib.Molecule, basis: veloxchem.veloxchemlib.MolecularBasis, origin: list[float] = [0.0, 0.0, 0.0]) -> list
Computes angular momentum integrals.
compute_linear_momentum_integrals(...) method of builtins.PyCapsule instance
compute_linear_momentum_integrals(molecule: veloxchem.veloxchemlib.Molecule, basis: veloxchem.veloxchemlib.MolecularBasis) -> list
Computes linear momentum integrals.
dipole_in_debye(...) method of builtins.PyCapsule instance
dipole_in_debye() -> float
Gets convertion factor for dipole moment (a.u. -> Debye).
extinction_coefficient_from_beta(...) method of builtins.PyCapsule instance
extinction_coefficient_from_beta() -> float
Gets factor needed for the calculation of the extinction coefficent from the electric-dipole magnetic-dipole polarizability beta.
fine_structure_constant(...) method of builtins.PyCapsule instance
fine_structure_constant() -> float
Gets fine-structure constant.
hartree_in_ev(...) method of builtins.PyCapsule instance
hartree_in_ev() -> float
Gets Hartree value in electronvolts.
hartree_in_kcalpermol(...) method of builtins.PyCapsule instance
hartree_in_kcalpermol() -> float
Gets Hartree value in kcal/mol.
hartree_in_kjpermol(...) method of builtins.PyCapsule instance
hartree_in_kjpermol() -> float
Gets Hartree value in kJ/mol.
hartree_in_wavenumber(...) method of builtins.PyCapsule instance
hartree_in_wavenumber() -> float
Gets Hartree value in reciprocal cm.
hartree_in_wavenumbers(...) method of builtins.PyCapsule instance
hartree_in_wavenumbers() -> float
Gets Hartree value in reciprocal cm.
is_chemical_element(...) method of builtins.PyCapsule instance
is_chemical_element(arg0: int) -> bool
Checks if identifier is chemical element number.
make_matrices(...) method of builtins.PyCapsule instance
make_matrices(*args, **kwargs)
Overloaded function.
1. make_matrices(arg0: int, arg1: CMolecularBasis, arg2: veloxchem.veloxchemlib.mat_t) -> CMatrices
Creates matrices for given basis.
2. make_matrices(arg0: Annotated[list[int], FixedSize(2)], arg1: CMolecularBasis, arg2: veloxchem.veloxchemlib.mat_t) -> CMatrices
Creates matrices for given basis.
3. make_matrices(arg0: Annotated[list[int], FixedSize(3)], arg1: CMolecularBasis, arg2: veloxchem.veloxchemlib.mat_t) -> CMatrices
Creates matrices for given basis.
4. make_matrices(arg0: Annotated[list[int], FixedSize(4)], arg1: CMolecularBasis, arg2: veloxchem.veloxchemlib.mat_t) -> CMatrices
Creates matrices for given basis.
5. make_matrices(arg0: int, arg1: CMolecularBasis, arg2: CMolecularBasis) -> CMatrices
Creates matrices for given basis.
6. make_matrices(arg0: Annotated[list[int], FixedSize(2)], arg1: CMolecularBasis, arg2: CMolecularBasis) -> CMatrices
Creates matrices for given basis.
7. make_matrices(arg0: Annotated[list[int], FixedSize(3)], arg1: CMolecularBasis, arg2: CMolecularBasis) -> CMatrices
Creates matrices for given basis.
8. make_matrices(arg0: Annotated[list[int], FixedSize(4)], arg1: CMolecularBasis, arg2: CMolecularBasis) -> CMatrices
Creates matrices for given basis.
make_matrix(...) method of builtins.PyCapsule instance
make_matrix(*args, **kwargs)
Overloaded function.
1. make_matrix(arg0: CMolecularBasis, arg1: veloxchem.veloxchemlib.mat_t) -> CMatrix
Creates matrix for given basis.
2. make_matrix(arg0: CMolecularBasis, arg1: CMolecularBasis) -> CMatrix
Creates matrix for given pair of bases.
mpi_master(...) method of builtins.PyCapsule instance
mpi_master() -> int
Gets the rank of MPI master process.
parse_xc_func(...) method of builtins.PyCapsule instance
parse_xc_func(xcLabel: str) -> veloxchem.veloxchemlib.XCFunctional
Converts exchange-correlation functional label to exchange-correlation functional object.
partition_atoms(...) method of builtins.PyCapsule instance
partition_atoms(arg0: int, arg1: int, arg2: int) -> list[int]
Get atomic indices of partitioned atoms list.
rotatory_strength_in_cgs(...) method of builtins.PyCapsule instance
rotatory_strength_in_cgs() -> float
Gets convertion factor for rotatory strength (a.u. -> 10^-40 cgs).
DATA
__commit__ = '0efbe13141bb0f03d3f33c374052bccf1086c552'
VERSION
1.0rc4
FILE
/Users/panor/miniconda3/envs/vlxman/lib/python3.12/site-packages/veloxchem/__init__.py
Classes#
The functionality of VeloxChem is built into the classes it implements. An overview of the available classes is provided with the dir function.
dir(vlx)
['AODensityMatrix',
'Absorption',
'AtomBasis',
'AtomBdeDriver',
'AtomTypeIdentifier',
'BasisFunction',
'BlockDavidsonSolver',
'BlockedGtoPairBlock',
'C6',
'C6Driver',
'CircularDichroismSpectrum',
'ComplexResponse',
'ConformerGenerator',
'CpcmDriver',
'CphfSolver',
'CubicGrid',
'CubicResponseDriver',
'DensityViewer',
'DensityViewerAlternative',
'DispersionModel',
'ElectricDipoleIntegralsDriver',
'ElectricDipoleMomentDriver',
'ElectricDipoleMomentGeom100Driver',
'EspChargesDriver',
'EvbDataProcessing',
'EvbDriver',
'EvbFepDriver',
'EvbForceGroup',
'EvbReporter',
'EvbSystemBuilder',
'ExcitedStateAnalysisDriver',
'ExcitonModelDriver',
'FirstOrderProperties',
'FockDriver',
'FockGeom1000Driver',
'FockGeom1010Driver',
'FockGeom1100Driver',
'FockGeom2000Driver',
'GradientDriver',
'GridDriver',
'GtoBlock',
'GtoPairBlock',
'HessianDriver',
'HessianOrbitalResponse',
'IMDatabasePointCollecter',
'IMForceFieldGenerator',
'InputParser',
'InterpolationDatapoint',
'InterpolationDriver',
'KineticEnergyDriver',
'KineticEnergyGeom100Driver',
'KineticEnergyGeom101Driver',
'KineticEnergyGeom200Driver',
'LinearAbsorptionCrossSection',
'LinearResponseEigenSolver',
'LinearResponseSolver',
'LinearResponseUnrestrictedEigenSolver',
'LinearResponseUnrestrictedSolver',
'LoPropDriver',
'MMDriver',
'MMForceFieldGenerator',
'MMGradientDriver',
'MOIntegralsDriver',
'Matrices',
'Matrix',
'MofBuilder',
'MolecularBasis',
'MolecularOrbitals',
'Molecule',
'Mp2Driver',
'MpiTask',
'NuclearPotentialDriver',
'NuclearPotentialErfDriver',
'NuclearPotentialErfGeom010Driver',
'NuclearPotentialErfGeom100Driver',
'NuclearPotentialGeom010Driver',
'NuclearPotentialGeom100Driver',
'NuclearPotentialGeom101Driver',
'NuclearPotentialGeom110Driver',
'NuclearPotentialGeom200Driver',
'NumerovDriver',
'OpenMMDriver',
'OpenMMDynamics',
'OpenMMGradientDriver',
'OptimizationDriver',
'OrbitalViewer',
'OutputStream',
'OverlapDriver',
'OverlapGeom100Driver',
'OverlapGeom101Driver',
'OverlapGeom200Driver',
'PEForceFieldGenerator',
'PolOrbitalResponse',
'Polarizability',
'PolarizabilityGradient',
'QuadraticResponseDriver',
'RIFockDriver',
'RIFockGradDriver',
'RIJKFockDriver',
'ReactionForceFieldBuilder',
'ReactionMatcher',
'RespChargesDriver',
'ResponseProperty',
'SHG',
'ScfGradientDriver',
'ScfHessianDriver',
'ScfRestrictedDriver',
'ScfRestrictedOpenDriver',
'ScfUnrestrictedDriver',
'Seminario',
'ShgDriver',
'SmdDriver',
'SolvationBuilder',
'SolvationFepDriver',
'SubCommunicators',
'SubMatrix',
'SymmetryAnalyzer',
'T3FlatBuffer',
'T4CScreener',
'TPA',
'TdaEigenSolver',
'TdaUnrestrictedEigenSolver',
'TddftGradientDriver',
'TddftOrbitalResponse',
'TdhfHessianDriver',
'ThreeCenterElectronRepulsionDriver',
'ThreeCenterElectronRepulsionGeom010Driver',
'ThreeCenterElectronRepulsionGeom100Driver',
'ThreeCenterOverlapDriver',
'ThreePATransitionDriver',
'TpaFullDriver',
'TpaReducedDriver',
'TpaTransitionDriver',
'TransitionStateGuesser',
'TwoCenterElectronRepulsionDriver',
'TwoCenterElectronRepulsionGeom100Driver',
'VibrationalAnalysis',
'VisualizationDriver',
'XCFunctional',
'XCIntegrator',
'XCMolecularGradient',
'XtbDriver',
'XtbGradientDriver',
'XtbHessianDriver',
'__builtins__',
'__cached__',
'__commit__',
'__doc__',
'__file__',
'__loader__',
'__name__',
'__package__',
'__path__',
'__spec__',
'__version__',
'aodensitymatrix',
'aoindices',
'assert_msg_critical',
'atombdedriver',
'atomtypeidentifier',
'available_functionals',
'available_pdft_functionals',
'batchsize',
'blockdavidson',
'bohr_in_angstrom',
'c2diis',
'c6driver',
'checkpoint',
'chemical_element_identifier',
'chemical_element_label',
'chemical_element_mass',
'chemical_element_max_angular_momentum',
'chemical_element_name',
'compute_angular_momentum_integrals',
'compute_electric_dipole_integrals',
'compute_kinetic_energy_integrals',
'compute_linear_momentum_integrals',
'compute_nuclear_potential_integrals',
'compute_overlap_integrals',
'configure_mkl_rt',
'conformergenerator',
'cpcmdriver',
'cphfsolver',
'cppsolver',
'cubicgrid',
'cubicresponsedriver',
'denmat',
'densityviewer',
'densityvieweralternative',
'dftutils',
'dipole_in_debye',
'dispersionmodel',
'distributedarray',
'doubleresbeta',
'environment',
'errorhandler',
'espchargesdriver',
'evbdataprocessing',
'evbdriver',
'evbfepdriver',
'evbreporter',
'evbsystembuilder',
'excitedstateanalysisdriver',
'excitondriver',
'extinction_coefficient_from_beta',
'features',
'fine_structure_constant',
'firstorderprop',
'fockdriver',
'get_basis_path',
'get_data_path',
'gradientdriver',
'griddriver',
'hartree_in_ev',
'hartree_in_kcalpermol',
'hartree_in_kjpermol',
'hartree_in_wavenumber',
'hartree_in_wavenumbers',
'hessiandriver',
'hessianorbitalresponse',
'imdatabasepointcollecter',
'imforcefieldgenerator',
'inputparser',
'interpolationdatapoint',
'interpolationdriver',
'is_chemical_element',
'linearsolver',
'lreigensolver',
'lreigensolverunrest',
'lrsolver',
'lrsolverunrest',
'make_matrices',
'make_matrix',
'mat_t',
'matrices',
'matrix',
'mklconf',
'mmdriver',
'mmforcefieldgenerator',
'mmgradientdriver',
'mofbuilder',
'mofmdprepare',
'mofoptimizer',
'mofpreparer',
'mofutils',
'mointsdriver',
'molecularbasis',
'molecularorbitals',
'molecule',
'molorb',
'mp2driver',
'mpi_master',
'mpitask',
'nonlinearsolver',
'numerovdriver',
'oneeints',
'openmmdriver',
'openmmdynamics',
'openmmgradientdriver',
'optimizationdriver',
'optimizationengine',
'orbitalviewer',
'outputstream',
'parse_xc_func',
'partition_atoms',
'peforcefieldgenerator',
'polarizabilitygradient',
'polorbitalresponse',
'print_features',
'profiler',
'pubchemfetcher',
'quadraticresponsedriver',
'reactionmatcher',
'read_results',
'reaffbuilder',
'respchargesdriver',
'rifockdriver',
'rijkfockdriver',
'rotatory_strength_in_cgs',
'rspabsorption',
'rspc6',
'rspcdspec',
'rsplinabscross',
'rsppolarizability',
'rspproperty',
'rspshg',
'rsptpa',
'sadguessdriver',
'sanitychecks',
'scfdriver',
'scfgradientdriver',
'scfhessiandriver',
'scfrestdriver',
'scfrestopendriver',
'scfunrestdriver',
'seminario',
'set_omp_num_threads',
'set_vlxbasispath',
'set_vlxdatapath',
'shgdriver',
'smddriver',
'smdsolventproperties',
'solvationbuilder',
'solvationfepdriver',
'subcommunicators',
'symmetryanalyzer',
'symmetryoperations',
'tdacppsolver',
'tdaeigensolver',
'tdaeigensolverunrest',
'tddftgradientdriver',
'tddftorbitalresponse',
'tdhfhessiandriver',
'threepatransitiondriver',
'tmparameters',
'tpadriver',
'tpafulldriver',
'tpareddriver',
'tpatransitiondriver',
'tsguesser',
'uffparameters',
'veloxchemlib',
'vibrationalanalysis',
'visualizationdriver',
'waterparameters',
'xtbdriver',
'xtbgradientdriver',
'xtbhessiandriver']
Information about a specific class is available from the docstring.
print(vlx.ScfUnrestrictedDriver.__doc__)
Implements spin unrestricted open shell SCF method with C2-DIIS and
two-level C2-DIIS convergence accelerators.
:param comm:
The MPI communicator.
:param ostream:
The output stream.
Methods and properties#
The actual calculations are performed by the methods in the classes. To list the properties and methods of a class, the dir function can be used as follows.
dir(vlx.ScfRestrictedDriver)
['__class__',
'__deepcopy__',
'__delattr__',
'__dict__',
'__dir__',
'__doc__',
'__eq__',
'__format__',
'__ge__',
'__getattribute__',
'__getstate__',
'__gt__',
'__hash__',
'__init__',
'__init_subclass__',
'__le__',
'__lt__',
'__module__',
'__ne__',
'__new__',
'__reduce__',
'__reduce_ex__',
'__repr__',
'__setattr__',
'__sizeof__',
'__str__',
'__subclasshook__',
'__weakref__',
'_apply_mom',
'_check_convergence',
'_comp_2e_fock',
'_comp_2e_fock_single_comm',
'_comp_density_change',
'_comp_diis',
'_comp_energy',
'_comp_full_fock',
'_comp_gradient',
'_comp_npot_mat_parallel',
'_comp_number_of_electrons',
'_comp_one_ints',
'_delete_mos',
'_gen_molecular_orbitals',
'_get_acc_type',
'_get_dyn_threshold',
'_get_effective_fock',
'_get_guess_type',
'_get_scaled_fock',
'_get_scf_range',
'_graceful_exit',
'_need_graceful_exit',
'_print_energy_components',
'_print_ground_state',
'_print_header',
'_print_iter_data',
'_print_scf_energy',
'_print_scf_finish',
'_print_scf_title',
'_store_diis_data',
'_update_mol_orbs_phase',
'_write_final_hdf5',
'comm',
'compute',
'compute_s2',
'density',
'gen_initial_density_proj',
'gen_initial_density_restart',
'gen_initial_density_sad',
'get_scf_energy',
'get_scf_type_str',
'history',
'is_converged',
'maximum_overlap',
'mol_orbs',
'molecular_orbitals',
'nnodes',
'nodes',
'num_iter',
'ostream',
'print_attributes',
'print_keywords',
'rank',
'scf_energy',
'scf_results',
'scf_tensors',
'scf_type',
'set_start_orbitals',
'update_settings',
'validate_checkpoint',
'write_checkpoint']
